MICROBIAL MOLECULAR BIOLOGY AND GENETICS
Microbial Recombination and Plasmids:
DNA Transformation:
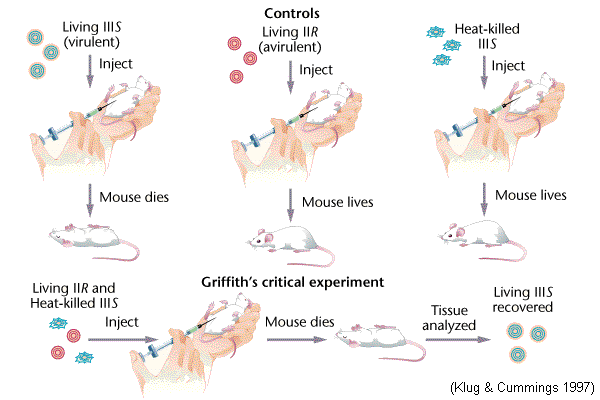
The second way in which DNA can move between bacteria is
through transformation, discovered by Fred Griffith in 1928.
Transformation is the uptake by a cell of a naked DNA molecule
or fragment from the medium and the incorporation of this molecule into the recipient chromosome in a heritable form. In natural transformation the DNA comes from a donor bacterium. The process is random, and any portion of a genome may be transferred between bacteria.
When bacteria lyse, they release considerable amounts of DNA
into the surrounding environment. These fragments may be relatively large and contain several genes. If a fragment contacts a competent cell, one able to take up DNA and be transformed, it can be
bound to the cell and taken inside. The transformation frequency of very competent cells is around 10^-3
for most genera when an excess of DNA is used. That is, about one cell in every
thousand will take up and integrate the gene. Competency is a complex phenomenon and is dependent on several conditions. Bacteria
need to be in a certain stage of growth; for example,S. pneumoniae
becomes competent during the exponential phase when the population reaches about 10^7 to 10
cells per ml. When a population becomes competent, bacteria such as pneumococci secrete a small
protein called the competence factor that stimulates the production, of 8 to 10 new proteins required for transformation. Natural transformation has been discovered so far only in certain gram-positive
and gram-negative genera: Streptococcus, Bacillus, Thermoactinomyces, Haemophilus, Neisseria, Moraxella, Acinetobacter,Azotobacter,and Pseudomonas.Other genera also may be capable
of transformation. Gene transfer by this process occurs in soil and
marine environments and may be an important route of genetic exchange in nature. The mechanism of transformation has been intensively studied in S. pneumoniae. A competent cell binds a
double-stranded DNA fragment if the fragment is moderately
large; the process is random, and donor fragments compete with
each other. The DNA then is cleaved by endonucleases to doublestranded fragments about 5 to 15 kilobases in size. DNA uptake
requires energy expenditure. One strand is hydrolyzed by an envelope-associated exonuclease during uptake; the other strand associates with small proteins and moves through the plasma membrane. The single-stranded fragment can then align with a
homologous region of the genome and be integrated. Transformation in Haemophilus influenzae,a gram-negative
bacterium, differs from that in S. pneumoniae in several respects.
Haemophilus does not produce a competence factor to stimulate
the development of competence, and it takes up DNA from only
closely related species (S. pneumoniae is less particular about the
source of its DNA). Double-stranded DNA, complexed with proteins, is taken in by membrane vesicles. The specificity of
Haemophilus transformation is due to a special 11 base pair sequence (5′AAGTGCGGTCA3′) that is repeated over 1,400 times
in H. influenzae DNA. DNA must have this sequence to be bound
by a competent cell.
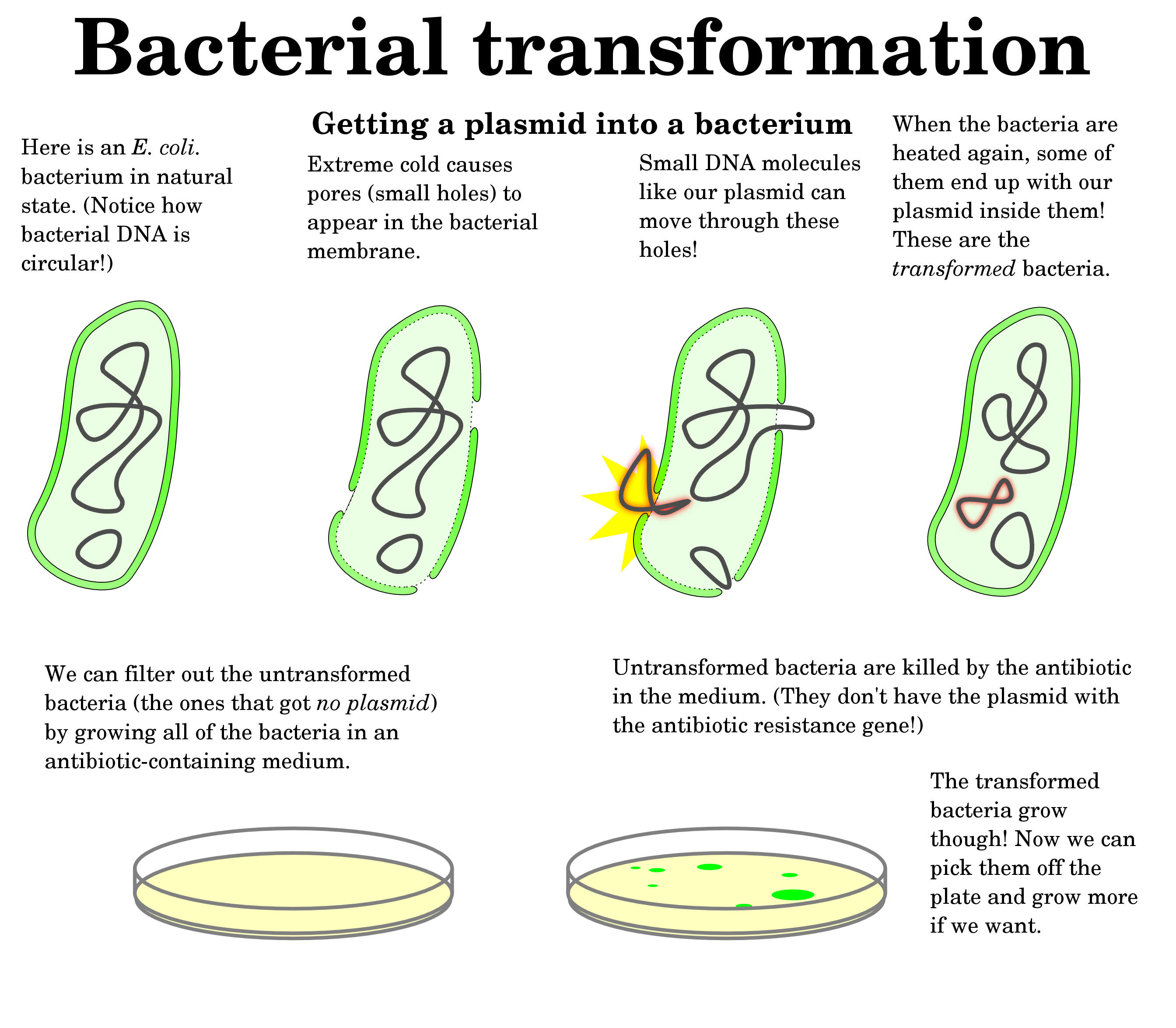
Artificial transformation is carried out in the laboratory by a
variety of techniques, including treatment of the cells with calcium chloride, which renders their membranes more permeable to DNA. his approach succeeds even with species that are not
naturally competent, such as E. coli.Relatively high concentrations of DNA, higher than would normally be present in nature,
are used to increase transformation frequency. When linear DNA
fragments are to be used in transformation,E. coli usually rendered deficient in one or more exonuclease activities to protect
the transforming fragments. It is even easier to transform bacteria with plasmid DNA since plasmids are not as easily degraded
as linear fragments and can replicate within the host.
Please refer this links too:
ecosal.org/mechanisms-of-dna-transformation.html
en.wikipedia.org/wiki/Transformation_(genetics)
www.ncbi.nlm.nih.gov/pubmed/18651316
www.ncbi.nlm.nih.gov/pubmed/21218625
web.biosci.utexas.edu/psaxena/.../050801DNA%20transformation.pdf
www.iisc.ernet.in/currsci/dec102002/1376.pdf
www.microbelibrary.org/.../3152-transformation-of-escherichia-coli-ma..
www.genome.ou.edu/protocol_book/protocol_adxF.html
Cited By Kamal Singh Khadka
Msc Microbiology, TU
Assistant Professor in Pokhara University, Regional College Of Science & Technology, PNC, LA, NA
Pokhara, Nepal.
DNA Transformation:
The second way in which DNA can move between bacteria is
through transformation, discovered by Fred Griffith in 1928.
Transformation is the uptake by a cell of a naked DNA molecule
or fragment from the medium and the incorporation of this molecule into the recipient chromosome in a heritable form. In natural transformation the DNA comes from a donor bacterium. The process is random, and any portion of a genome may be transferred between bacteria.
When bacteria lyse, they release considerable amounts of DNA
into the surrounding environment. These fragments may be relatively large and contain several genes. If a fragment contacts a competent cell, one able to take up DNA and be transformed, it can be
bound to the cell and taken inside. The transformation frequency of very competent cells is around 10^-3
for most genera when an excess of DNA is used. That is, about one cell in every
thousand will take up and integrate the gene. Competency is a complex phenomenon and is dependent on several conditions. Bacteria
need to be in a certain stage of growth; for example,S. pneumoniae
becomes competent during the exponential phase when the population reaches about 10^7 to 10
cells per ml. When a population becomes competent, bacteria such as pneumococci secrete a small
protein called the competence factor that stimulates the production, of 8 to 10 new proteins required for transformation. Natural transformation has been discovered so far only in certain gram-positive
and gram-negative genera: Streptococcus, Bacillus, Thermoactinomyces, Haemophilus, Neisseria, Moraxella, Acinetobacter,Azotobacter,and Pseudomonas.Other genera also may be capable
of transformation. Gene transfer by this process occurs in soil and
marine environments and may be an important route of genetic exchange in nature. The mechanism of transformation has been intensively studied in S. pneumoniae. A competent cell binds a
double-stranded DNA fragment if the fragment is moderately
large; the process is random, and donor fragments compete with
each other. The DNA then is cleaved by endonucleases to doublestranded fragments about 5 to 15 kilobases in size. DNA uptake
requires energy expenditure. One strand is hydrolyzed by an envelope-associated exonuclease during uptake; the other strand associates with small proteins and moves through the plasma membrane. The single-stranded fragment can then align with a
homologous region of the genome and be integrated. Transformation in Haemophilus influenzae,a gram-negative
bacterium, differs from that in S. pneumoniae in several respects.
Haemophilus does not produce a competence factor to stimulate
the development of competence, and it takes up DNA from only
closely related species (S. pneumoniae is less particular about the
source of its DNA). Double-stranded DNA, complexed with proteins, is taken in by membrane vesicles. The specificity of
Haemophilus transformation is due to a special 11 base pair sequence (5′AAGTGCGGTCA3′) that is repeated over 1,400 times
in H. influenzae DNA. DNA must have this sequence to be bound
by a competent cell.
Artificial transformation is carried out in the laboratory by a
variety of techniques, including treatment of the cells with calcium chloride, which renders their membranes more permeable to DNA. his approach succeeds even with species that are not
naturally competent, such as E. coli.Relatively high concentrations of DNA, higher than would normally be present in nature,
are used to increase transformation frequency. When linear DNA
fragments are to be used in transformation,E. coli usually rendered deficient in one or more exonuclease activities to protect
the transforming fragments. It is even easier to transform bacteria with plasmid DNA since plasmids are not as easily degraded
as linear fragments and can replicate within the host.
Please refer this links too:
ecosal.org/mechanisms-of-dna-transformation.html
en.wikipedia.org/wiki/Transformation_(genetics)
www.ncbi.nlm.nih.gov/pubmed/18651316
www.ncbi.nlm.nih.gov/pubmed/21218625
web.biosci.utexas.edu/psaxena/.../050801DNA%20transformation.pdf
www.iisc.ernet.in/currsci/dec102002/1376.pdf
www.microbelibrary.org/.../3152-transformation-of-escherichia-coli-ma..
www.genome.ou.edu/protocol_book/protocol_adxF.html
Cited By Kamal Singh Khadka
Msc Microbiology, TU
Assistant Professor in Pokhara University, Regional College Of Science & Technology, PNC, LA, NA
Pokhara, Nepal.



Comments